coppafish
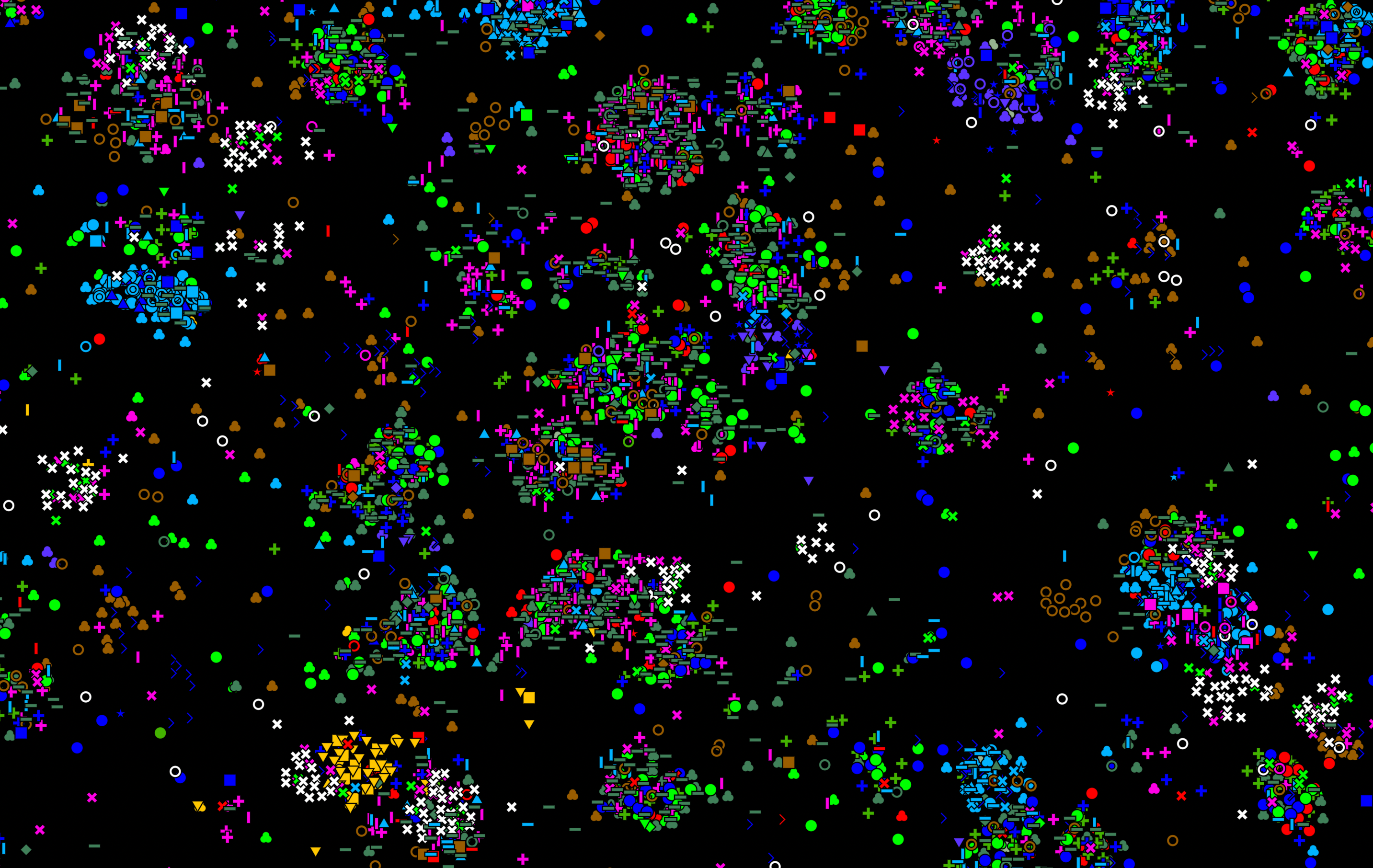
coppafish is a data analysis pipeline for decoding coppaFISH (combinatorial padlock-probe-amplified fluorescence in situ hybridization) datasets. coppaFISH is a method for in situ transcriptomics which produces a series of images arranged in terms of tiles, rounds and channels. coppafish then determines the distribution of genes via image processing, spot detection, registration and gene calling. The distribution of genes (like that shown above - each marker refers to a different gene) can then be used to determine the location and type of cells using pciSeq.
Installation
coppafish supports python 3.8 and above. It can be installed using pip:
pip install coppafish
To use the napari Viewer and matplotlib diagnostics, the plotting version must be installed:
pip install coppafish[plotting]
To use the optimised code, which is recommended for running the find spots and OMP sections of the pipeline (otherwise they are very slow), the optimised version must be installed:
pip install coppafish[optimised]
Installing on Windows
The optimised code requires jax which is not supported on Windows, thus the optimised version of coppafish cannot be used on Windows.
The optimised and plotting features can both be installed by running:
pip install coppafish[optimised,plotting]